[ad_1]
After gaining notoriety for its superhuman performance in game simulations, artificial intelligence group DeepMind has solved a scientific problem that eluded researchers for more than 50 years.
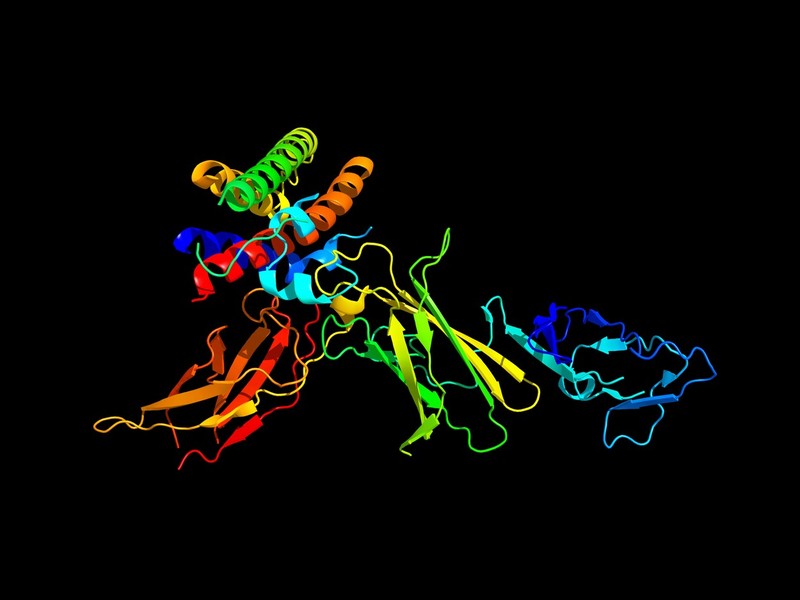
AlphaFold, the latest artificial intelligence program developed by the company, has shown that it can predict how proteins adapt to 3D shapes – an extraordinarily complex but fundamental process for understanding the biology of life.
Independent researchers say the discovery will help researchers understand the mechanisms behind the disease and pave the way for certain specialty drugs, more nutritious crops and “green enzymes” that break down plastic.
“This is an exciting time for the industry,” said Demis Hassabis, founder and CEO of DeepMind.
“These algorithms are becoming mature and powerful enough to be applied to truly challenging scientific questions,” he added.
Venki Ramakrishnan, president of the Royal Society of Great Britain, described DeepMind’s work as “surprising progress” that occurred “decades earlier than people on the ground would have expected.”
Understanding how proteins are positioned has been a challenge for researchers over the past 50 years, as they are a kind of molecular origami whose importance is difficult to stress.
Most biological processes revolve around proteins, and the shape of a protein determines its function.
When researchers know how a protein is deposited, they can begin to find out what it does. How insulin regulates blood sugar or how antibodies fight coronavirus is determined by the structure of the proteins.
Scientists have identified over 200 million proteins, but the structures are known for only a fraction of them.
Traditionally, shapes are discovered after meticulous laboratory studies which can take years to be extremely complex work.
Proteins are chains of amino acids that can twist and fold into a surprising number of shapes: a googol in a cube, i.e. a 1 followed by 300 zeros.
To learn how to position proteins, DeepMind researchers trained their algorithm using a public database containing 170,000 sequences and shapes of proteins.
The training lasted several weeks, DeepMind used the equivalent of 100-200 graphics processing units – modest processing power by modern standards,
The AlphaFold algorithm not only outperformed other programs, but achieved a level of accuracy comparable to meticulous and time-consuming laboratory methods.
Hassabis said DeepMind is already working with scientists to give them access to the AlphaFold program and help them with their research work.
Andrei Lupas, director of the Department of Developmental Biology at the Max Planck Institute in Germany, said he has already used the program to solve a protein-related problem that the researchers failed to clarify for a decade.
Read also:
.
[ad_2]
Source link