[ad_1]
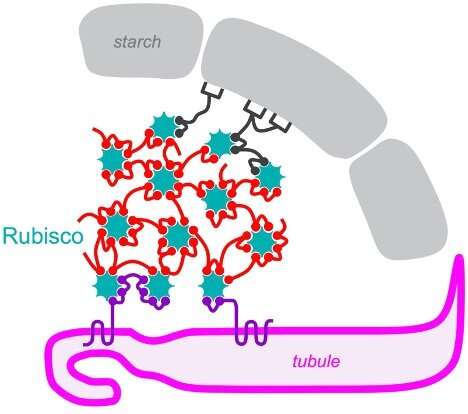
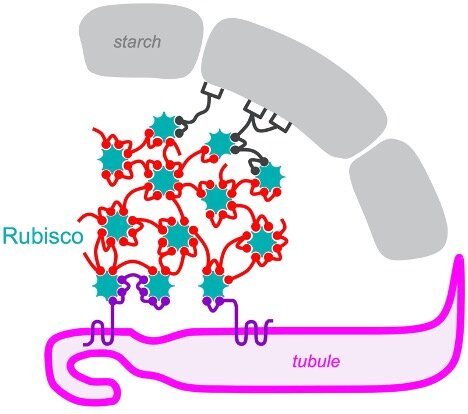
Princeton researchers led by Moritz Meyer and Martin Jonikas found that the algal pyrenoid, which concentrates carbon dioxide for the Rubisco enzyme, self-assembles through interactions between Rubisco and other pyrenoid proteins containing a pattern (sphere shapes in the diagram). Credit: Meyer et al.
Next time you visit a lake or beach, take a deep breath. As you exhale, take a moment to be thankful for the little things: specifically, for the microscopic unicellular algae in the soil and waters all around you that are extracting the carbon dioxide you just exhaled and incorporating it into the sugars that eventually they will be used by every other organism in the biosphere. About 30% of this activity, globally, is carried out by a specialized algae facility called a pyrenoid.
To visualize a pyrenoid, think of a pomegranate. The pyrenoid contains Rubisco grains, the enzyme that does the molecular work of incorporating carbon dioxide into sugars. These grains are embedded in a support pulp, or matrix, of other proteins, which is itself surrounded by an outer shell made of starch. The fruit is somewhat eaten by worms; it is riddled with finger-like channels – actually, membrane-enclosed tubules – that supply concentrated carbon dioxide to the Rubisco beans. Tubules are important for pyrenoid function because waterborne algae such as Chlamydomonas reinhardtii would otherwise have difficulty obtaining enough carbon dioxide to keep Rubisco running at full capacity.
The pyrenoid presents several puzzles for scientists. For example, how the proteins that make up the pyrenoid are addressed there and how they organize themselves into such a complex arrangement has been an enduring mystery. The new lab work of Martin Jonikas, assistant professor at the Princeton Department of Molecular Biology and collaborators, has now solved this conundrum.
“The main initial discovery was made by accident,” says Jonikas.
Molecular research biologist Moritz Meyer and colleagues were trying to identify which proteins are present in the pyrenoid besides Rubisco. To do this, they used an antibody – a protein that, like a key, binds to other proteins that possess a specific and corresponding block. Meyer and colleagues planned to break the algae open and then add an antibody that binds a particular matrix protein to the resulting molecular soup. By pulling the antibody, the scientists could extract that protein. Any other protein that binds to the antibody’s target protein would have come for the ride, and the scientists could then determine if any of them were a previously unknown pyrenoid component. But the experiment didn’t go as planned.
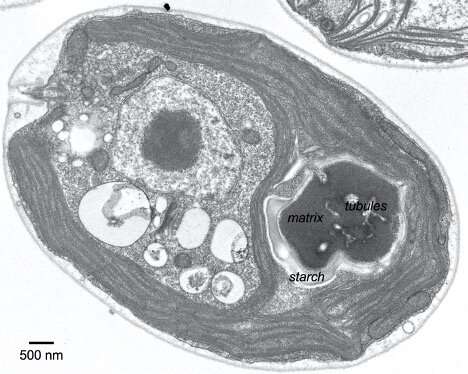
Princeton researchers found that the algal pyrenoid, which concentrates carbon dioxide for the Rubisco enzyme, self-assembles through interactions between Rubisco and other pyrenoid proteins containing a pattern. Here, an electron micrograph of an algal cell, with parts of the pyrenoid labeled. Credit: Meyer et al.
“We noticed that the antibody binds directly to several proteins located in the pyrenoids,” says Jonikas. In other words, they had just discovered that all of these proteins have a lock that matches the key to their antibody. Closer examination of the proteins revealed the existence of an amino acid sequence, or motif, which is present in the original antibody target and also appears in all other proteins.
“We hypothesized that this motif could serve as a signal that directs proteins to the pyrenoid, and the experiments we have done support this hypothesis,” explains Jonikas. “Removing the motif from one of the proteins containing the motif caused it to no longer localize in the pyrenoid, while adding it to non-pyrenoid proteins caused it to localize the pyrenoid.”
Meyer and colleagues found that the motif is linked to Rubisco. This explains how the pyrenoid is formed: its component proteins remain free in the cell until they collide with Rubisco and become trapped.
“Many of the proteins do not simply locate on the pyrenoid matrix, but rather appear to locate on the interfaces between the matrix and the other two pyrenoid subcompartments, the pyrenoid tubules and the starch sheath,” Jonikas notes. This can allow proteins to self-organize into the complex pyrenoid structure.
“The study represents an exquisite example of investigative science,” says Dr Howard Griffiths, professor of plant science at the University of Cambridge in the UK. Dr. Griffiths collaborated with Jonikas’ group on other studies but was not involved in this work.
“They used clever experimental manipulations to show that a common motif could allow the specific linker to form the Rubisco matrix and anchor other key elements both internally to the thylakoid tubules and the starch sheath towards the periphery,” Griffiths says. “Overall, the report by Meyer and colleagues made a significant contribution to our understanding of the shape and function of pyrenoids, with relevance both for understanding aquatic primary productivity and for supporting approaches seeking to incorporate such a mechanism for ‘turbocompress’ photosynthesis in land crops plants. ”
Researchers explore how an organelle is formed that fixes carbon via phase separation
Moritz T. Meyer et al, the assembly of the CO2-fixing algal organelle, the pyrenoid, is driven by a Rubisco binding motif, Advances in science (2020). DOI: 10.1126 / sciadv.abd2408
Provided by Princeton University
Quote: Scientists Discover Motive Driving Algal Pyrenoid Assembly (2020, November 25) Retrieved November 25, 2020 from https://phys.org/news/2020-11-scientists-motif-algal-pyrenoid.html
This document is subject to copyright. Aside from any conduct that is correct for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.
[ad_2]
Source link