
[ad_1]
Any biological sample, such as dirt, water or food, contains billions of bacteria. Only a few are harmful to humans or pathogenic. But those few pathogens can make the difference between a reliable supply of meat or lettuce, for example, and a food poisoning epidemic or, worse, a pandemic.
Discovering deadly pathogens before they strike is one of the goals of the Defense Advanced Research Projects Agency (DARPA). The agency, part of the United States Department of Defense, works on developing new technologies to help protect the United States.
A few years ago, DARPA launched a “Friend or Foe” challenge to the research community to see if they could quickly isolate pathogens from a very complicated sample without affecting the sample’s basic functionality or phenotype. One of the three institutions that took up the challenge, Pacific Northwest National Laboratory (PNNL) formed a multidisciplinary team with scientific expertise in soil microbiology and proteomics, biochemical synthesis and data analysis.
Now, halfway through a project that started in 2019, the team is perfecting a promising new method developed for pathogen discovery.
The method, called OmniScreen, is an end-to-end pipeline to quickly and effectively distinguish a plethora of pathogenic cells in a microbial community. The system extracts, probes and screens thousands of cells for pathogens within days.
Notably, OmniScreen also keeps samples alive.
Typically, in biological research, a sample is frozen and then “fixed” as needed for use. Fixing solution can kill bacteria or at least alter their original phenotype. Overcoming this hurdle was crucial for PNNL to get the green light from DARPA for the next phase of their project.
“We weren’t really sure it would work. Part of the risk we identified early on was that the cells might not survive the survey, “said Becky Hess, a biomedical scientist at PNNL and project leader for OmniScreen.” This was one of the criteria for moving to phase two and. ‘we made it work “.
Priming the bacteria tubing

The first challenge was to extract the bacteria hidden in tiny pockets of a soil sample. Led by microbiologist and lab colleague Janet Jansson, the team tried different centrifugation techniques and established density gradients. After the sample was centrifuged in a neutral solution, the heavy soil particles settled to the bottom and the lighter bacteria floated to the top.
The technique initially produced one million cells in three hours, two hours longer than the team’s initial goal, which they continue to work towards. Jansson stressed the need for speed in extraction and analytical techniques for DARPA.
Subsequently, Jansson separated the living cells from the dead ones and stained them to facilitate identification during subsequent phenotyping. This proteomics analysis took place at the Environmental Molecular Sciences Laboratory (EMSL), a US Department of Energy Office of Science user facility located on the PNNL-Richland campus. EMSL’s powerful mass spectrometers mapped the protein-associated traits in each cell.
Comparisons of subsamples before and after the proteomics step confirmed that the phenotypes still matched. To keep the cells alive and the phenotype of each cell intact, Jansson came up with a simple yet innovative soil maintenance tea.
“When we immerse the soil in water for the spin phase, we end up with some kind of tea,” explained Jansson. “It works perfectly to prevent cells from going into shock in a new environment.”
Priming and breaking down pathogens
Bacteria are a type of microbe that may contain specific characteristics, or chemistry, that make them pathogenic. Wright uses information on microbial characteristics to build chemical probes with similar molecular traits. The probes serve as a bait to catch bacteria with an affinity for the same traits.
“Like a fish on a hook, once the microbe grabs a probe, that’s it, it’s stuck,” said Aaron Wright, a biomedical scientist in NLP’s Division of Biological Sciences. Wright added that the different colors on the probes give them away. “We know what fish we caught.”
A reporter mechanism is also integrated into each probe for monitoring purposes. In this case, that mechanism is fluorescence. Once a probe attaches to a protein, it emits light of a certain color depending on the pathogen.
Wright then uses a flow cytometer to sort the bacteria one cell at a time at a rate of over 10 million cells per hour. The instrument can tell if the cell has a colored probe, and therefore pathogenic characteristics (or not), and inserts them accordingly.
“We can create all kinds of lures to hook different types of pathogens,” Wright said.
In the laboratory, the technique is called multiplexing; multiple probes can rapidly and simultaneously affect multiple pathogenic traits.
Disease Cell Screening
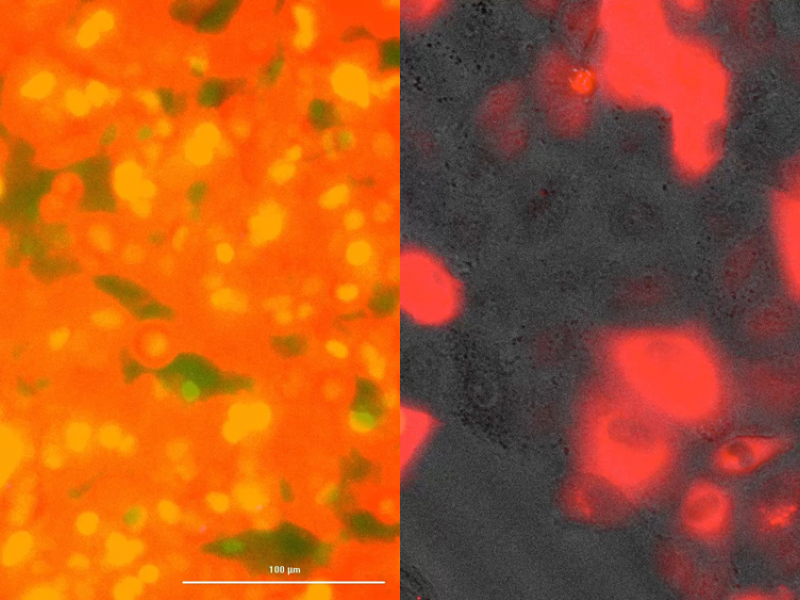
Once explored, the bacteria are found in a plastic petri dish along with the human lung, intestine and immune cells. The team uses lung and intestinal cells because they are the most common routes for infection in humans. Under a standard microscope, healthy growing cells are seen to adhere strongly to the plate, but diseased cells lose structural integrity and begin to lift off the plate.
“Pathogens can evade immune cells and cause damage to lung or intestinal tissues,” Hess said. “When cell junctions begin to fall apart, it’s an indication of exposure to pathogens.”
Damaged cells also glow.
“This is our validation phase,” Hess said. “If human cells appear healthy, there are no pathogens present. If human cells lose structural integrity or shine, this tells us that a pathogen is being introduced. “
But that validation is a laborious and time-consuming step. Hess said one test could produce 15,000 images. Even with an automated imaging microscope, it took days to determine pathogenicity by manually examining all of the images. This hurdle is where machine learning, the third segment of the pipeline, comes into play.
Machine learning accelerates the discovery of pathogens
Enoch Yeung, assistant professor at the University of California, Santa Barbara, developed the machine learning algorithm to recognize cellular characteristics that indicate sick cells versus healthy cells.
Yeung conceived the OmniScreen idea for the “Friend or Foe” challenge while working on other research as a visiting scientist in the National Security Directorate of PNNL. Aware of Hess’s experience in immunology and synthetic biology, he asked her, “Can you have human cells tell you if they’re sick?” He said, “Yes, I can.”
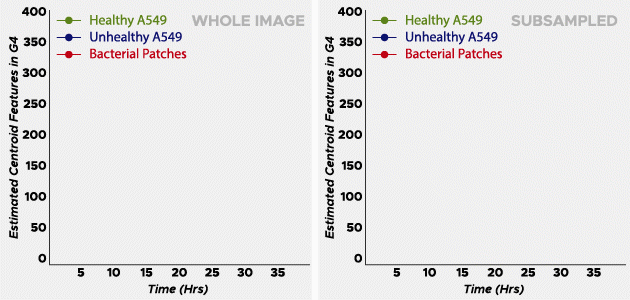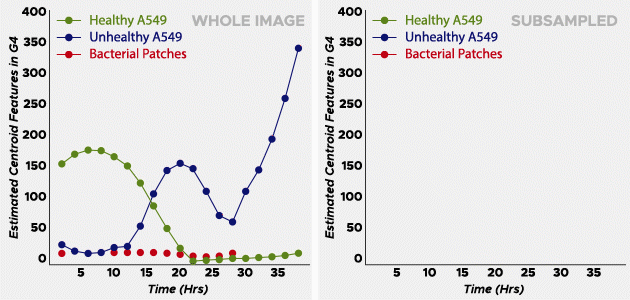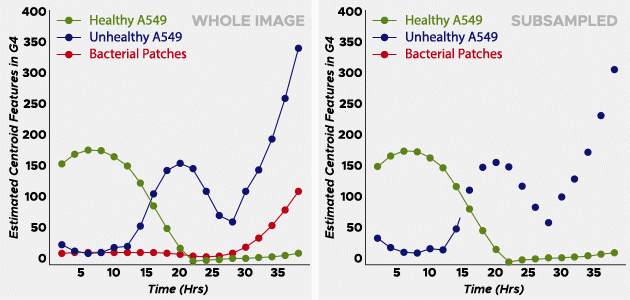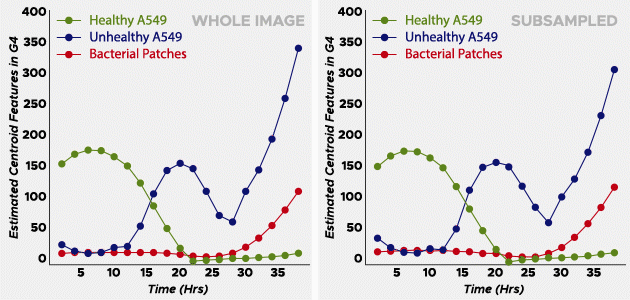
The two scientists examined his collection of healthy and unhealthy cells to create a sparse and labeled dataset to train the algorithm. Learning from those images and the color, or phenotype, of the pathogen, the algorithm quickly screened thousands of images and generated data graphs with three S-shaped curves: healthy cells crashing (dying), unhealthy cells into increase and growth rate of bacteria colonies. The steeper the curve on the left side of the plot, the more aggressive the pathogen is.
Increase productivity
In phase one of the tests, the OmniScreen algorithm resolved 30 species of bacteria with 92% accuracy in one week. The basic truth sample provided by DARPA contained 19 pathogenic bacteria; OmniScreen performed 17 – much better than expected.
Entering phase two of the $ 8.4 million project, the team’s next challenge is to solve a sample containing nearly twice the number of bacteria than the soil sample, but from an unknown source.
As in phase one, Robert Egbert, a synthetic biologist, focuses on addressing the critical integration of tools and process steps across the system.
“We are continually working on handoffs between technical areas,” said Hess, “but having a blind sample is a real test of the system.”
.
[ad_2]
Source link