[ad_1]
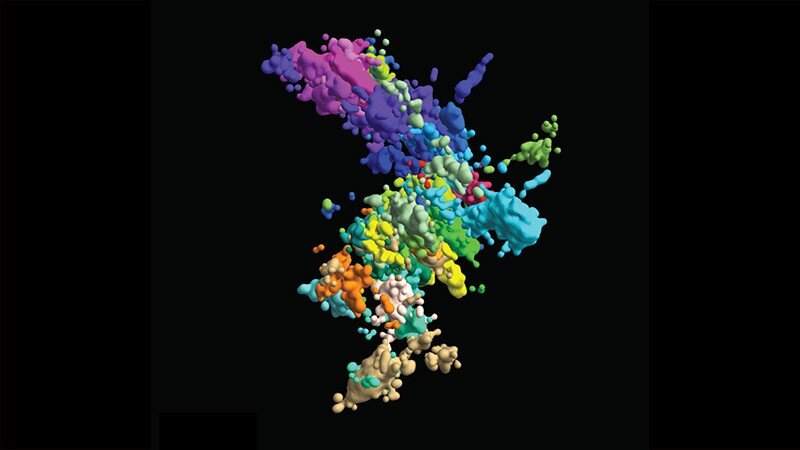
This multicolored chromatin image was created using multiplex fluorescence in situ hybridization and super resolution microscopy. Credit: Xiaowei Zhuang laboratory
In high school textbooks, human chromosomes are depicted as wobbly Xs like two hot dogs squeezed together. But those images are far from accurate. “90 percent of the time,” Jun-Han Su said, “chromosomes don’t exist like that.”
Last year, before Su graduated with his Ph.D., he and three current PhD students. Graduate School of Arts and Sciences candidates – Pu Zheng, Seon Kinrot, and Bogdan Bintu – captured high-resolution 3-D images of human chromosomes, the complex houses of our DNA. Now, those images could provide enough evidence to turn those Xs into more complex but far more accurate symbols not only to teach the next generation of scientists, but also to help the current generation unravel the mysteries of how chromosome structure affects functioning.
All living things, including humans, must create new cells to replace those that are too old and worn out to function. To do this, cells divide and replicate their DNA, which is wrapped in labyrinthine libraries within chromatin, the substance within chromosomes. Stretched in a straight line, the DNA in a single cell can reach six feet, all wrapped up in tight and complex structures in a cell’s nucleus. A single mistake while copying or rewinding genetic material could cause genes to mutate or malfunction.
Zooming in enough to see the chromatin structure is difficult. But looking at both structure and function is even more difficult. Now, in an article published in August in Cell, Zhuang and his team report on a new method for visualizing chromatin structure and behavior together, connecting dots to determine how one affects the other to maintain proper function or cause disease.
“It is very important to determine the 3-D organization,” said Zhuang, David B. Arnold, Jr. Professor of Science, “to understand the molecular mechanisms underlying the organization and also to understand how this organization regulates the function of the genome “.
With their new high-resolution 3-D imaging method, the team began building a chromosome map from both wide-angle images of all 46 chromosomes and close-ups of a section of a chromosome. To visualize something that is still too small to visualize, they captured linked points (“genomic loci”) along each DNA chain. By connecting many dots, they could form a complete picture of the chromatin structure.
But there was a hitch. Previously, Zhuang said, the number of points they could represent and identify was limited by the number of colors they could represent together – three. Three dots cannot create a complete image.
So, Zhuang and his team came up with a sequential approach: image three different locations, turn off the signal, and then imagine three more in quick succession. With this technique, each point gets two identifying marks: color and round image.
“We now actually have 60 loci simultaneously captured and located and, importantly, identified,” Zhuang said.
However, to cover the entire genome, they needed more, thousands, so they turned to a language already used to organize and store huge amounts of information: binary. By imprinting binary barcodes on different chromatin loci, they could visualize many more places and decode their identities at a later time. For example, a molecule that was imaged in the first round but not in the second receives a barcode starting with “10.” With 20-bit barcodes, the team could differentiate 2,000 molecules in just 20 imaging rounds. “In this combinatorial way, we can increase the number of molecules that are visualized and identified much faster,” said Zhuang.
With this technique, the team imaged around 2,000 chromatin loci per cell, a more than ten-fold increase from previous work and enough to form a high-resolution image of what the structure of chromosomes looks like in its natural habitat. But they didn’t stop there: they also envisioned transcription activity (when RNA replicates genetic material from DNA) and nuclear structures such as nuclear stains and nucleoli.
With their 3-D maps of the Google genome, they could begin to analyze how the structure changes over time and how those territorial movements help or damage cell division and replication.
Researchers already know that chromatin is broken down into different areas and domains (like deserts versus cities). But what those media look like in different cell types and how they work is still unknown. With their high-resolution images, Zhuang and the team determined that areas with many genes (“rich in genes”) tend to cluster in similar areas on any chromosome. But areas with few genes (“poor in genes”) only join if they share the same chromosome. One theory is that gene-rich areas, which are active sites for gene transcription, come together like a factory to allow for more efficient production.
Although more research is needed before confirming this theory, one thing is now certain: the local chromatin environment affects transcription activity. Structure influences function. The team also found that no two chromosomes are alike, even in otherwise identical cells. Finding out what every chromosome looks like in every cell in the human body will take a lot more work than a laboratory can do alone.
“It will not be possible to build on our work alone,” Zhuang said. “We have to rely on the work of many, many labs to have a complete understanding.”
Scientists identify “dumbbell-like” structures of DNA that encode proteins
Jun-Han Su et al, Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin, Cell (2020). DOI: 10.1016 / j.cell.2020.07.032
Cell
Provided by Harvard University
Quote: Chromosomes Look Different Than You Think (2020, Nov 18) retrieved Nov 18, 2020 from https://phys.org/news/2020-11-chromosomes.html
This document is subject to copyright. Aside from any conduct that is correct for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.
[ad_2]
Source link