[ad_1]
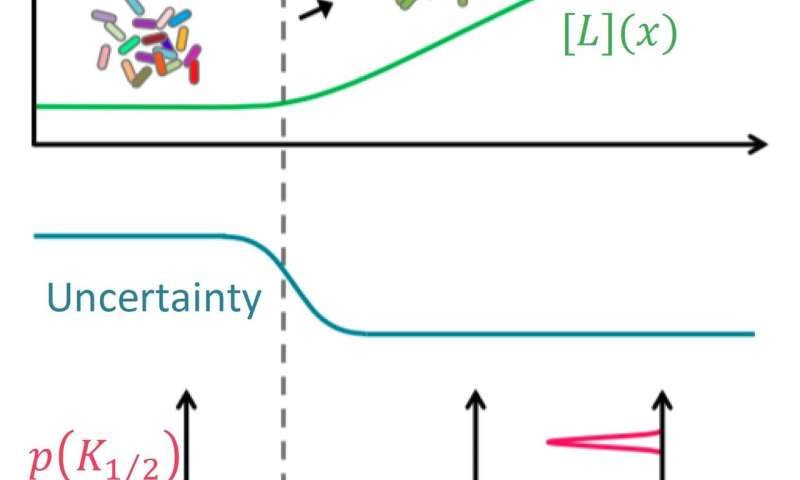
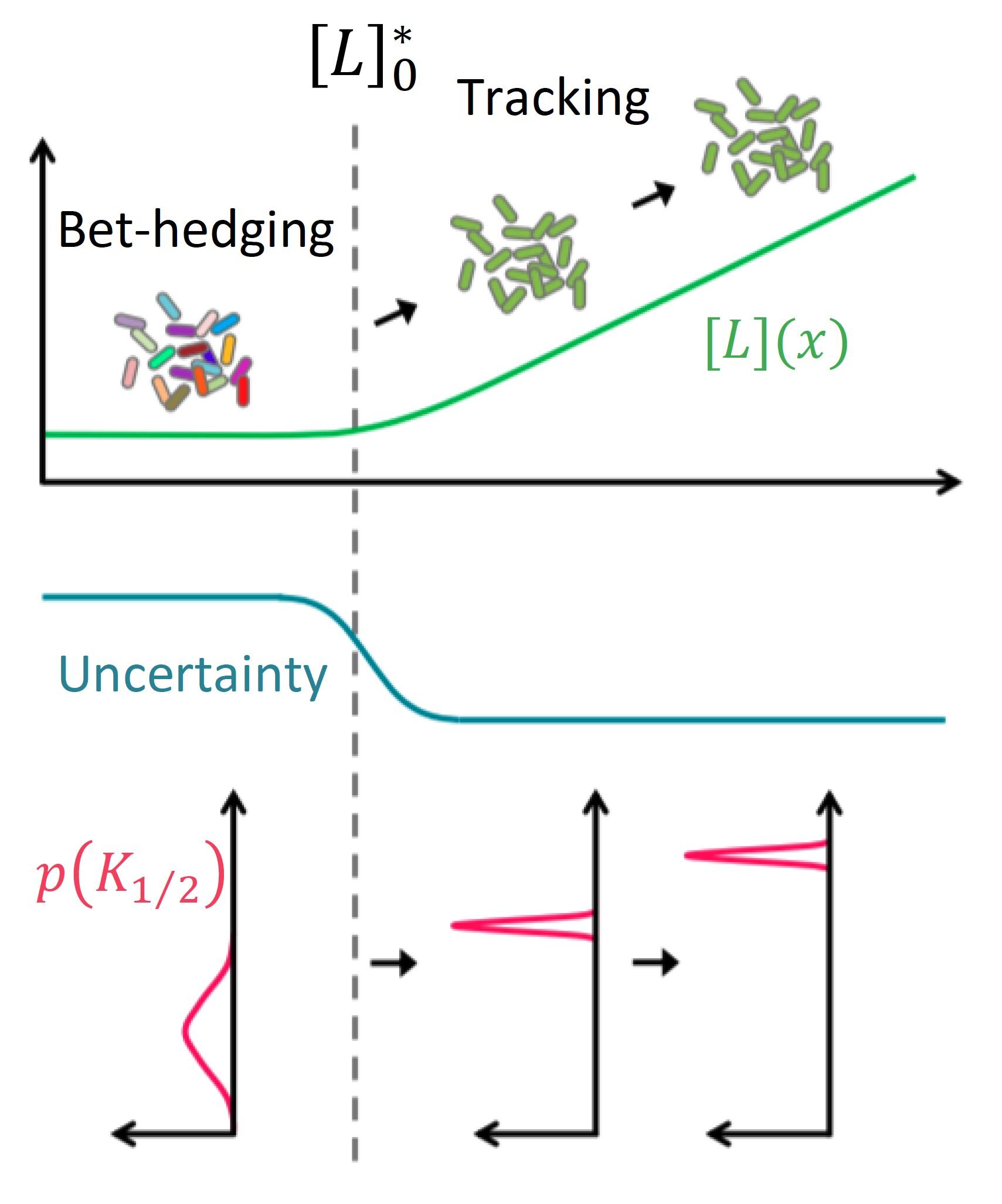
Dynamic risk management by cell populations. In the absence of environmental cues, uncertainty about the environment is high (middle panel) and the E. coli cell population was found to exhibit strong diversity in their sensory response (indicated by the wide distribution of the response coefficient K_ (1/2) in the lower panel), allowing responses to many different signals (schematized by the cartoon color of the cells inside the upper panel in the “betting coverage” scheme). But once a given signal [L] (green curve in the top panel) has exceeded a threshold (〖[L]〗 _0 ^ *) predicted by the mathematical model, sensory diversity has collapsed (indicated by the narrow distribution of K_ (1/2) in the lower panel), allowing the entire population to ‘focus’ on that particular signal (‘ tracking ‘in the upper panel). Credit: K. Kamino
Just like investors in the stock market, cell populations prepare for changes in the environment by spreading risk. The toolbox they use contains a repertoire of sensory receptors on the surface of individual cells. These receptors can be modified to make individual members of the population sensitive to different environmental signals. It was thought that cells could change this diversity only relatively slowly, either by producing new receptor proteins or by degrading them. Scientists from AMOLF (Amsterdam, Netherlands) and Yale University (New Haven, CT) now report the discovery of a mechanism that allows cell populations to tune their diversity much faster, through a combination of physical and chemical interactions between existing proteins. The results are published in the journal Advances in science on November 13.
The new experimental findings reveal that E. coli bacterial cell populations retain high diversity in their sensory portfolio when environmental signals are absent, but this diversity dramatically decreases (by a factor of 10) when subjected to new environmental signals. “This makes a lot of sense,” says Tom Shimizu, a group leader at AMOLF and the study’s senior author. “When environmental signals are scarce, uncertainty about the future is at its peak, so the smart thing to do is spread your bets widely on many possible new signals. But the game changes when you sense a certain stimulus. Evidently, cell populations. they use that new information to “focus” their attention on a particular signal, so that the entire population can respond together. ”
What was surprising, however, was how quickly the cells could switch to this new strategy. In biological cells, up until now it was assumed that updates to the sensory capability portfolio required changes in gene expression, a time-consuming process involving the synthesis of new protein molecules or the degradation of old ones. “However, these cell populations could adjust how they spread their bets on different sensory modalities in seconds,” says Keita Kamino, lead author of the study who initiated the study at AMOLF and finished it at Yale. “This immediately ruled out the possibility that the cells were updating their bets via gene expression, which takes minutes or hours.”
To study the mechanism at work, the team then built a mathematical model of observed diversity, based on known interactions between receptors and other molecules that process and transmit signals across the cell. While the model does not include a way for cells to modify their gene expression, they found that it very accurately predicts the changes in sensory diversity of cells seen in previous experiments. “Surprisingly, diversity tuning emerges from the combination of allosteric interactions and covalent modifications between protein molecules,” says Thierry Emonet, professor at Yale University and corresponding co-author of the study, referring to the physical influence of proteins in contact between they cause changes in their shape (known as “allosteric interactions”) and the addition and removal of chemical groups at those same receptors (known as “covalent modification”). “Although a model capturing these interactions had already been established by a large body of experimental work, the consequences of this allosteria for diversity attunement were not appreciated.”
Therefore, the team identified a simple explanation for the rapid changes in risk spreading achieved by the cell population. Instead of building and breaking down protein molecules, the mechanism works through chemical modifications of existing proteins and their effects on the physical cross-talk between those proteins, allowing for much faster movement. Such protein modifications are widely used to process and transmit signals within single cells, but it was previously not known that they could also play a role in the diversification of entire cell populations. Because such changes are so prevalent in all cells of all organisms, the authors believe that this underlying mechanism for rapid changes in cell diversity may be at work in many cell types throughout biology.
The virus that causes COVID-19 blocks cellular defenses
K. Kamino, JM Keegstra, J. Long, T. Emonet, TS Shimizu, Adaptive tuning of cellular sensory diversity without changes in gene expression, Advances in science, advances.sciencemag.org/lookup… .1126 / sciadv.abc1087
Quote: Dynamic Risk Management in Cell Populations (2020, November 13) Retrieved November 13, 2020 from https://phys.org/news/2020-11-dynamic-cell-populations.html
This document is subject to copyright. Apart from any conduct that is correct for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.
[ad_2]
Source link