[ad_1]
Just as biologists draw evolutionary trees whose branches represent the development of different species and the relationships between them, some scientists now look to cells to draw another type of family tree. This tree describes not the millions of species that inhabit our planet, but the millions of neurons that make up the kumquat-sized brain of a laboratory mouse.
Before the advent of DNA sequencing, scientists used visual cues to define species. Does the animal have fur or feathers? A beak, claws, hooves? The same goes for early brain studies: Over the past 100 years, neuroscientists have come to realize that our neurons and other brain cells have a wide variety of shapes and sizes. But detailed visual descriptions of these cells have failed to reveal many insights into how different neurons do different jobs, let alone how individual cells might change in disease.
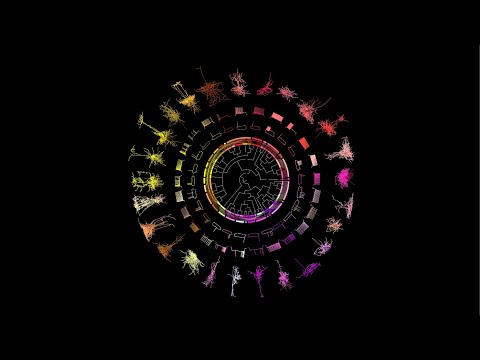
The Allen Institute team used a special technique to simultaneously capture the 3D shape of a brain cell, its unique electrical properties, and the suite of genes it activates, from hundreds of individual neurons on the part of the mouse brain that processes information. visual. Because this technique captures multiple types of data from each individual cell, researchers can use this data set to classify neurons into different types, part of their larger effort to build a “periodic table” of the mammalian brain. The data and the cell classification scheme can link together other studies that focus on a single characteristic, resulting in a more holistic understanding of different brain cells and their relationships.
By classifying brain cells into categories or cell types and studying how and where these cell types do their work, researchers hope to understand more about how the brain as a whole works and where it could go wrong in disease.
“It is becoming increasingly clear that in some disorders, there is a deficiency in a very particular type of neuron in a particular part of the brain,” he said. Gabe Murphy, Ph.D., Associate Director of Electrophysiology at the Allen Institute for Brain Science. “The more we understand the different types of neurons that exist and what makes them unique, the more we can understand what goes wrong if you are vulnerable to one or more of those types of disease.”
Murphy conducted the study together with the Allen Institute for Brain Science’s Hongkui Zeng, Ph.D., Executive Vice President and Director; Nathan Gouwens, Ph.D., Assistant Investigator; is Staci Sorensen, Ph.D., Associate Director of Neuroanatomy.
“This study essentially provided a ‘lookup table’ for other neuroscientists, so that if you have information on just one property of a neuron, you can deduce the other properties,” he said. Edward Callaway, Ph.D., Professor at the Salk Institute for Biological Studies and advisor to the Allen Institute for Brain Science, who was not involved in the study. “We need this kind of linking study to understand what the true types of brain cells are so that we can begin characterizing them. This dataset enables that linking and will undoubtedly prove to be a great resource for future discovery.”
Researchers at the Allen Institute focused on inhibitory neurons, the type of cells that restrain the activity of other neurons. Their study divided these neurons into 28 different types. Scientists had not previously realized that this level of diversity existed in visual inhibitory neurons.
Revealing new patterns in the brain
Neuroscientists use characteristics such as the shape of a neuron, the number and type of connections it makes with other neurons, the types of electrical signals it sends and receives, and the genes it turns on, to sort cells into cell types. Most studies focus on just one of these attributes. The Allen Institute researchers used a technique known as Patch-seq to capture three from a single cell: the 3D shape of a neuron, or morphology; its unique electrical properties, or electrophysiology, which researchers record by “applying” a glass electrode to the cell; and the gene sequences it activates, or its transcriptomic profile.
Patch-seq is a complex technique. To generate this large data set, the Allen Institute team created a “pipeline” to ensure that the results were standardized and of high quality. The team acquired gene and electrical expression data from over 4,200 neurons in the current study. As creating complete 3D morphologies takes a long time, they focused on 517 neurons to track. This phase was performed with input from players playing the free neuroscience game Mozak, developed by researchers from the Allen Institute and the University of Washington Center for Game Science.
It’s a 21st turning point on a branch of neuroscience dating back to 19thscientists of the century who first described many neurons based on their shape.
“With this approach, we are now learning something new about the brain by adding transcriptomics to the method of studying neuronal morphology that has been around for more than 100 years,” Sorensen said. “The use of transcriptomics to label cell types has been revolutionary. When we sort cells in this way, we are able to reveal patterns we haven’t noticed before.”
In the short term, Allen Institute researchers want to use their neuron lookup table to reveal more information about an ongoing project to map the brain’s electrical pattern. Researchers in this IARPA-supported collaboration are creating images and mapping one cubic millimeter of a mouse’s brain in extraordinary detail. The 3D shapes of the brain cells present in that tiny brain cube can be mapped to Patch-seq data, allowing scientists to gather new insights into how brain cell types connect to form circuits.
The research described in this article was partially supported by awards from the National Institutes of Health, including the National Eye Institute award numbers R01EY023173 and the National Institute of Mental Health U01MH105982 and Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is the sole responsibility of the authors and does not necessarily represent the official views of NIH and its subsidiaries.
For a complete list of authors, see the study. Hongkui Zeng and Zizhen Yao of the Allen Institute are also co-authors of a complementary study published online today in the journal Nature, which uses Patch-seq to describe the diversity of neurons in the mouse motor cortex.
Information on the Allen Institute for Brain Science
The Allen Institute for Brain Science is a division of the Allen Institute (alleninstitute.org), an independent 501 (c) (3) nonprofit medical research organization, and is dedicated to accelerating understanding of how the brain human functioning in health and pathology. Using a great scientific approach, the Allen Institute generates useful public resources used by researchers and organizations around the world, drives technological and analytical advances, and uncovers the fundamental properties of the brain through the integration of experiments, models and theory. Launched in 2003 with an initial contribution from founder and philanthropist, the late Paul G. Allen, the Allen Institute is supported by a variety of government, foundation and private funds to enable its projects. Data and tools from the Allen Institute for Brain Science are publicly available online at brain-map.org.
Media contact:
Rob Piercy, Director, Media Relations
206.548.8486 | [email protected]
SOURCE Allen Institute for Brain Science

Related links
http://www.alleninstitute.org
.
[ad_2]
Source link