[ad_1]
The 2019 coronavirus disease (COVID-19) pandemic calls for better and faster tests to ensure more accurate diagnosis, monitoring and surveillance of the disease and its spread. Serological testing is used to identify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection after the acute phase.
A new study published on the prepress server bioRxiv* in November 2020 describes the virus’s N protein as a promising epitope for serology.
Antibody response in COVID-19
Seroprevalence studies, which measure the presence of a pathogen in a population through blood tests, can be used to define appropriate policies in the management of the COVID-19 outbreak. However, this depends on the sensitivity and accuracy of the test itself. Currently, the available tests are associated with a high percentage of false positives and false negatives.
Most serological studies conducted so far have described the initiation of the antibody response to SARS-CoV-2, targeting the spike (S) and nucleocapsid (N) protein antigens, starting on the fourth day following the first symptom. At this point, specific IgM and IgG antibodies are detected, simultaneously or one after the other.
It is generally observed that the antibody titers stabilize around the sixth day after the first antibody detection. At present, most immunoassays focus on recombinant S and N antigens. The S antigen is exposed on the viral surface, while the N protein is the most abundant viral protein during infection.

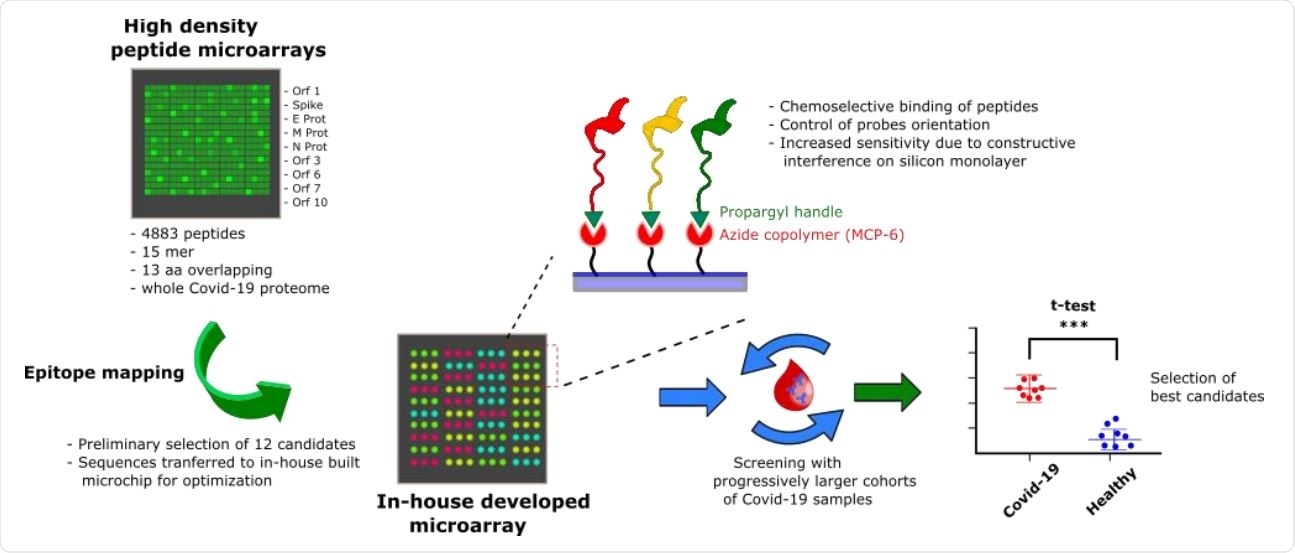
Probe selection workflow overview. A high-density peptide microarray showing the entire SARS-CoV-2 proteome was probed for immunoreactivity with COVID-19 serum samples, leading to the selection of 12 peptide shots. The most promising candidates were then transferred to a low-density microarray platform for site-oriented, selective peptide immobilization and validated with progressively larger and independent patient cohorts to select the most sensitive and specific immunoreactive peptide probes.
Problems with full-length antigens
However, full-length recombinant antigens have several disadvantages. They are expensive, difficult to store and subject to deterioration over time and environmental fluctuations. Quality can therefore vary greatly between batches. Additionally, cross-reaction potential is another issue that can confuse the final test result.
This is especially true of SARS-CoV-2 as some of the viral proteins are closely related to those expressed by phylogenetically close human coronaviruses, which are also commonly found in the human population at the same time, such as those that cause the common cold.
Even the very regions of these viral proteins targeted by virus-specific antibodies are not fully described at the moment, although full understanding of this could help explain different disease outcomes. It could also help in the development of several immunology-based tests and therapies.

(A): SARS-CoV 2 Spike protein (S) with its 12 domains (NTD: n-terminal domain, S1 and S2: furin cleavage site, CD: connection domain, HR2: heptad repeat 2, FP: fusion peptide, CH: central helix). The epitopes are highlighted as follows: AM55 in red; AM50 in yellow; AM64 in purple; AM 63 in blue, AM 49 in green. (B): SARS-CoV 2 nucleocapsid (N) protein, N-terminal and C-terminal domains. The epitopes are highlighted as follows: AM57 in green, AM66 in blue. (C): AM57 sequence alignment on SARS CoV 2, SARS CoV, MERS CoV, hCoV-OC43.
Advantages of synthetic peptides
The use of synthetic peptide probes is a promising solution to these problems as they are stable for storage, economically manufactured, and have high batch-to-batch consistency. Furthermore, problems with the secondary and tertiary structure of proteins no longer arise. The ability to synthesize peptides in the exact sequence needed makes them highly versatile so they can be used in a wide range of diagnostic settings, whether for point-of-care testing, lateral flow testing or for centralized laboratory workflows using ELISA and bead-mounted test.
Selection and validation of epitopes
To help understand antigen-antibody interactions in primary antigenic regions, current researchers used peptide microarrays. This platform allows multiplex screening of up to thousands of peptides simultaneously. The peptides are immobilized like spots on a solid support, arranged according to ordered patterns.
The researchers used more than 4,800 peptides covering the entire viral proteome, from serum samples from seven PCR-confirmed, antibody-positive COVID-19 elderly patients. All had a history of mild or moderate disease approximately one month before serum collection. Each peptide was 15 amino acids long, with 13 overlapping amino acids.
They found 12 possible linear epitopes on the N and S protein and the ORF1ab polyprotein. Using the microarray technology they had already developed, which uses site-selectively arranged and oriented peptides, they validated the 12 epitopes with serum from another 12 patients, with the same time frame, as well as serum collected before the first diagnosis 19, as of December 2018. Of the 12, six epitopes could discriminate between cases and controls.
Increasing the number of test samples to 28, collected from patients 1-5 months after symptom onset, reduced the hit epitopes to 3, one each from the N and S protein and one from the ORF1ab polyprotein. Of these, the epitope in protein N was highly immunoreactive and was specific in its binding to antibodies from infected individuals compared to negligible binding in control sera.
High discriminating power for N-Epitope AM57
The researchers concluded that protein N had the linear epitope (AM57) with the best specificity and sensitivity, in the region 155-171. Furthermore, this epitope shows only 53% homology with the N protein of seasonal coronaviruses.
They validated their conclusion using 50 samples from other COVID-19 patients, collected 1-5 months after the first symptom. This demonstrated that the anti-AM57 IgG antibodies showed a high discriminating power, with a sensitivity of 92% and a specificity of 100%. This is very close to the performance offered by the full N antigen.
IgM antibodies against this epitope also had a high discriminating power, but not against the full-length N antigen. The lack of false negatives with IgG antibodies could be attributed to the lack of preservation of this epitope among other coronaviruses, thus preventing pre-existing cross-reactivity.
In addition, the AM57 epitope was analyzed using 5 longitudinal series of sera collected over months from disease onset. The researchers found that within two weeks of infection, as defined by a positive PCR test, both IgM and IgG were detectable. The IgG antibody response but not the IgM response persisted for the 7 weeks of testing.
Implications
The study concludes: “The ability of the AM57 peptide to detect the IgM response early deserves further investigation.” Furthermore, the authors state: “Synthetic peptides offer clear advantages in terms of cost and synthetic versatility by allowing for simple implementation in different diagnostic settings: from lateral flow tests for point of need diagnostics to ELISA and bead based assays. for centralized settings “.
The ability of peptide microarrays to process large numbers of peptides or samples simultaneously could potentially allow the formation of more specific synthetic probes for the diagnosis of COVID-19. Another promising branch could be the identification of novel targets for the immunotherapy of the infection.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and therefore should not be considered conclusive, guide clinical practice / health-related behavior, or treated as consolidated information.
.
[ad_2]
Source link