[ad_1]
A long-standing and incredibly complex scientific problem regarding the structure and behavior of proteins has been effectively solved by a new artificial intelligence (AI) system, the scientists report.
DeepMind, the UK-based artificial intelligence company, has been thrilling us for years with its parade of ever-changing neural networks that continually defeat humans in complex games like chess and Go.
However, all of these incremental advances were more than just mastering recreational diversions.
In the background, DeepMind researchers were trying to persuade their AIs to solve far more important scientific puzzles, such as finding new ways to fight disease by predicting infinitesimal but vital aspects of human biology.
Now, with the latest version of their AlphaFold AI engine, they seem to have actually achieved this very ambitious goal, or at least have brought us closer than scientists have ever done before.
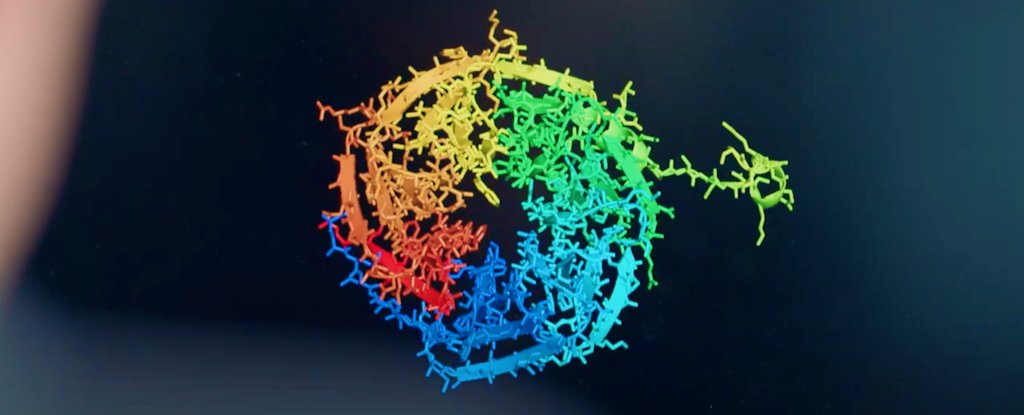
For nearly 50 years, researchers have struggled to predict how proteins achieve their three-dimensional structure, and it’s not an easy problem to solve.
The astronomical number of potential configurations is so staggering, in fact, that the researchers speculated that it would take longer than the age of the Universe to sample all possible molecular arrangements.
However, if we can solve this conundrum, known as the protein folding problem, it would be a huge step forward in scientific capabilities, greatly accelerating research efforts in things like drug discovery and disease modeling and also leading to new applications. far beyond health.
For this reason, despite the scale of the challenge, researchers have collaborated for decades to achieve improvements in developing solutions to the problem of protein folding.
A rigorous experiment called CASP (Critical Assessment of protein Structure Prediction) began in the 1990s, challenging scientists to devise systems that can predict the esoteric riddles of protein folding.
Now, in its third decade, the CASP experiment appears to have produced its most promising solution – with DeepMind’s AlphaFold providing predictions of 3D protein structures with unprecedented accuracy.
“We’ve been stuck on this problem – how proteins fold – for nearly 50 years,” says CASP co-founder John Moult of the University of Maryland.
“To see DeepMind produce a solution for this, after working on this problem personally for so long and after so many stops and you start wondering if we would ever get there, is a very special moment.”
In the experiment, DeepMind used a new deep learning architecture for AlphaFold that can interpret and compute the “spatial graph” of 3D proteins, predicting the molecular structure underlying their folded configuration.
The system, which was trained by analyzing a database of approximately 170,000 protein structures, brought its unique skill set to this year’s CASP challenge, called CASP14, achieving an average score in its predictions of 92.4 GDT (Global Distance Test).
It’s above the ~ 90 GDT threshold which is generally considered competitive with the same results obtained via experimental methods, and DeepMind says its predictions are only about 1.6 angstroms on average (about the width of an atom).
“I nearly fell off my chair when I saw these results,” says genomics researcher Ewan Birney of the European Molecular Biology Laboratory.
“I know how rigorous CASP is: it basically ensures that computational modeling has to work in the challenging task of from the beginning protein folding. It was humbling to see that these models could do it so accurately. There will be many things to understand, but this is a huge advance for science. “
It is worth noting that the research has not yet been peer-reviewed, nor published in a scientific journal (although the DeepMind researchers say it is on the way).
Even so, experts familiar with the field are already acknowledging and applauding the breakthrough, though the full report and detailed results have yet to be seen.
“This computational work represents an extraordinary advance on the problem of protein folding, a great 50-year challenge in biology,” says structural biologist Venki Ramakrishnan, president of the Royal Society.
“It happened decades before many people in the field had predicted.”
The full results have not yet been published, but you can see the abstract for the research, “High Precision Protein Structure Prediction Using Deep Learning”, here, and find more information on CASP14 here.
.
[ad_2]
Source link