[ad_1]
In a recent medRxiv * preprint manuscript, researchers from Singapore and the United States report the development of a digital CRISPR method for the sensitive, specific and rapid detection of viral nucleic acids at constant temperature, which could be exploited for the detection of severe acute respiratory syndrome coronavirus 2 (SARS- CoV-2).
To combat the global spread of coronavirus disease (COVID-19), improved methods are urgently needed to detect SARS-CoV-2 and quantify viral load. Although reverse transcription polymerase chain reaction (RT-qPCR) is a current gold standard, quantification requires standards or external references with often variable results.
This is where digital PCR comes in, with highly sensitive detection and viral load analysis of SARS-CoV-2; however, the reaction time is extended and takes about four hours compared to qPCR, which takes one hour.
Likewise, CRISPR-based methods can also detect SARS-CoV-2 within an hour but do not allow for absolute quantification of viral particles, which would reduce inter-laboratory variability and further open the scientific field.
This research effort, led by Dr. Xiaolin Wu of the Singapore-MIT Alliance for Research and Technology, reveals an optimized RApid DIgital Crispr Approach (RADICA) that enables absolute quantification of viral nucleic acids at a constant temperature in one hour.

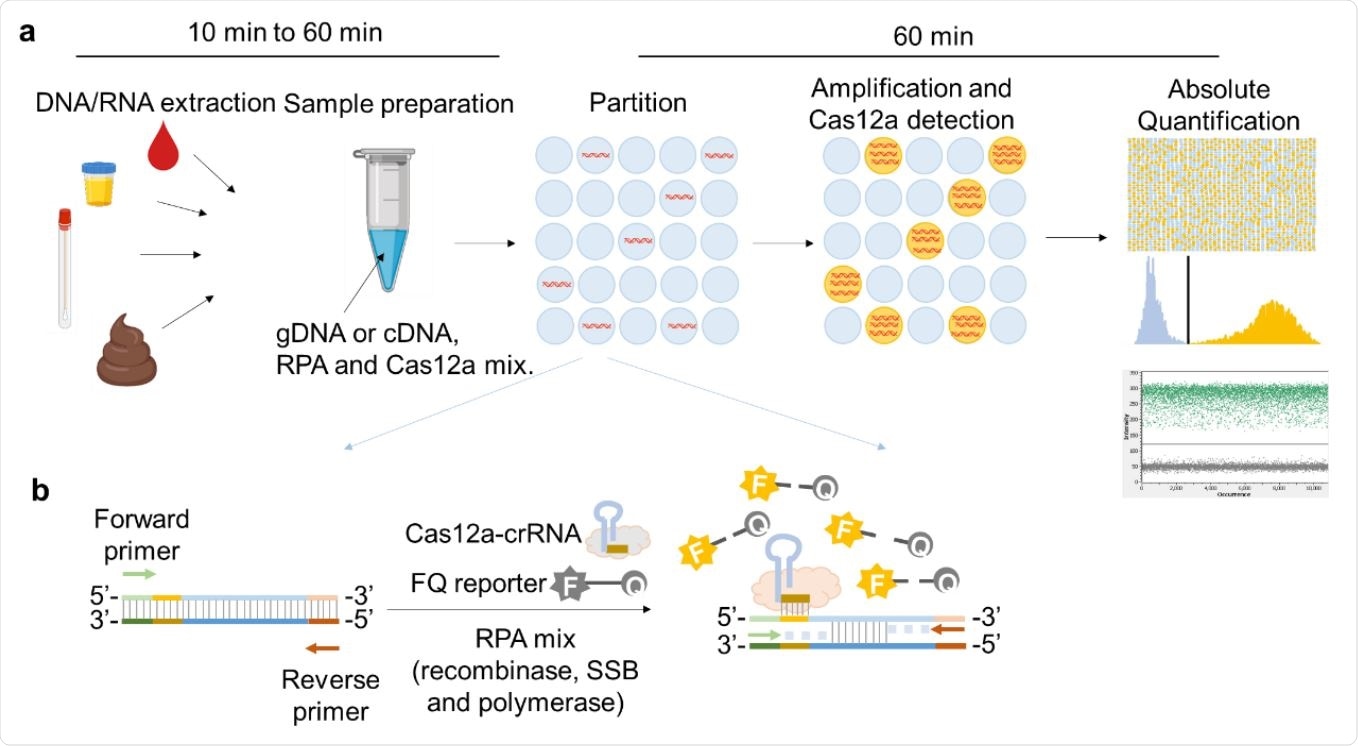
Schematic illustration of RADICA. a, The RADICA sample partitioning workflow on a chip for absolute quantification of nucleic acid targets. Generally, after the DNA / RNA extraction step, different types of clinical samples can be used for the detection and quantification of various targets. The sample mix containing DNA / cDNA, RPA reagents and Cas12a-crRNA715 FQ probes is randomly distributed across thousands of partitions. In each partition, DNA is amplified by RPA and detected by Cas12a-crRNA, resulting in a fluorescent signal in the partition. Based on the proportion of positive partitions and the Poisson distribution, the absolute copy number of the nucleic acid target is quantified. b, Illustration of the RPA-Cas12a reaction in each positive partition. In each partition containing the target nucleic acid, the primers bind to the target nucleic acid and initiate amplification with the help of recombinase and DNA polymerase. Due to the DNA polymerase strand displacement, the exposed crRNA-targeted ssDNA sites are linked by Cas12a-crRNA complexes. Cas12a is then activated and cleaves the neighboring FQ reporters to produce a fluorescence reading.
Optimized vessel reaction on a high-density commercial chip
The method presented in this paper combines all the advantages of quantitative digital PCR, rapid isothermal amplification, and specific CRISPR detection in a single vessel reaction system that separates individual reactions into ten thousand compartments on a high-density commercial chip.
Such a simplified one-vessel reaction combines CRISPR-based detection and nucleic acid amplification in a single step in a closed tube, significantly reducing the risk of sample cross-contamination during batch processing.
For validation purposes, DNA containing the SARS-CoV-2 nucleoprotein gene was used, which showed a linear signal-to-input response. Furthermore, RADICA was compared with digital PCR to quantify synthetic SARS-CoV-2 DNA and Epstein-Barr viral DNA (EBV).
The researchers also tested the possible inhibitory effects of the underlying DNA on the reactions that were carried out in small partitions. The isothermal feature of the RADICA-based detection test uses a simple constant temperature heat bath, which can allow for rapid viral detection that can be used even in areas with few resources.
Aggravating effect of accuracy and speed
In this study, the use of the RADICA method led to sensitivity and detection limits comparable to those of real-time PCR and other isothermal methods, with absolute quantification capability. Furthermore, a significant advantage of RADICA observed in this study was its speed.
The above absolute DNA quantification showed a dynamic range from 0.6 to 2027 copies per microliter, with no cross-reactivity on similar viruses or human background DNA. The results of this study also suggest that the background DNA will not inhibit the reaction of the RADICA sample within the dynamic range that will be used for testing purposes.
Additionally, when it comes to speed, RADICA can accurately detect and quantify nucleic acid in one hour without thermal cycling, an alternative four times faster than digital PCR-based virus detection.
Potential uses of innovative technology
“We established and characterized RADICA, which combines the speed and sensitivity of isothermal amplification, the specificity of CRISPR-based detection, and the ability to achieve absolute quantification through sample partitioning,” the study authors summarize their methodological breakthrough.
The absolute quantification approach for viral detection may not only support a simpler clinical process, but could also be exploited for research purposes. And since viral loads can be linked to transmission risk and disease severity (as in the case of SARS-CoV-2), this could also prove crucial in addressing this pandemic.
Future work will focus on expanding RADICA’s applications to scientific areas such as gene expression analysis, rare mutant detection, sequencing library quantification and copy number variation, with significant benefits for industries. clinical, pharmaceutical, ecological and public health.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behaviors, or treated as consolidated information.
.
[ad_2]
Source link